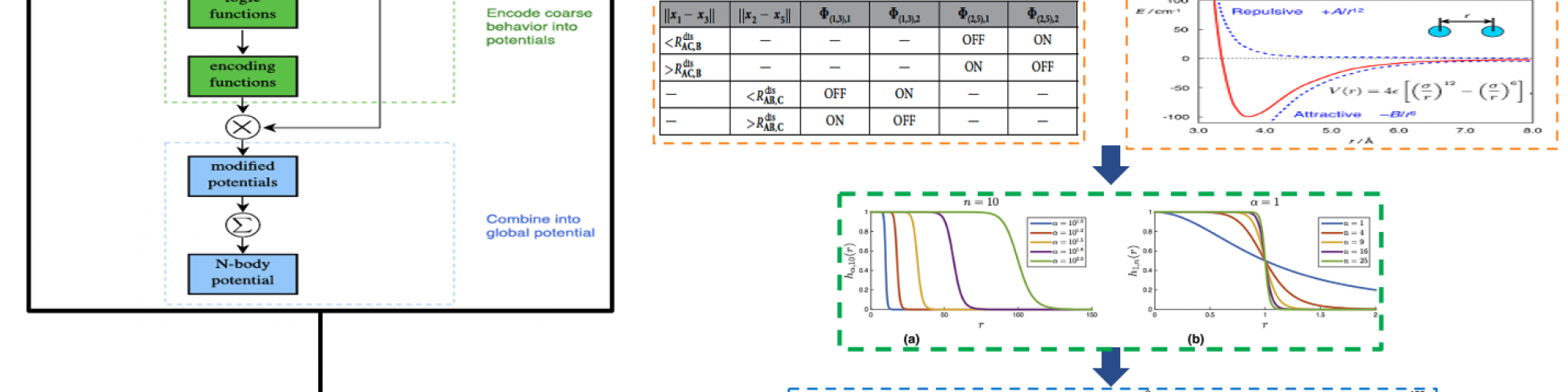
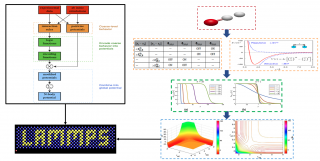
At the highest level, quantum-accurate training data is generated for the reactions of interest. Then a standard "off the shelf" two-body potential is selected. Next the coarse level logic that describes the intended reaction is established and the form of the encoding functions necessary to enforce the logic is derived. The parameters of the potential and encoding functions are obtained via a least square regression over the training data. The two-body potentials are then "stitched" together via encoding function multiplication to establish a multi-body reactive potential. Finally, the multi-body programmable potential is imputed into the molecular simulation package LAAMPS to verify the potential produces the intended reaction and thus generalizes beyond the training data set.
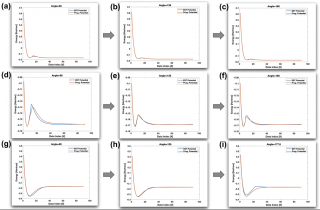
We compare our results for a chain reaction that starts with H_2+O ---> H+OH and proceeds to form a water molecule via H+OH ---> H2O. The data is collected across various fixed O-H-H angles of approach while varying the O-H_a and H_a-H_b distances.
Top Row: Lammps Simulation for 2H+O ---> H+OH. Bottom Row: Lammps Simulation for OH+H ---> H2O.